Clustering
Clustering
- Broad set of techniques for finding subgroups.
- Observations within groups are quite similar to each other and observations in different groups quite different from each other.
Clustering methods
- Partitioning: k-means clustering
- Agglomerative/divisive: hierarchical clustering
- Model-based: mixture models
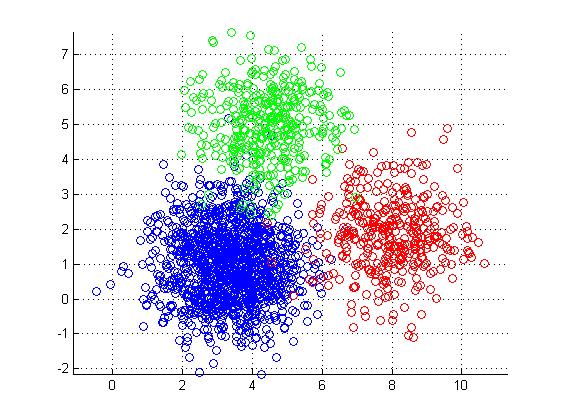
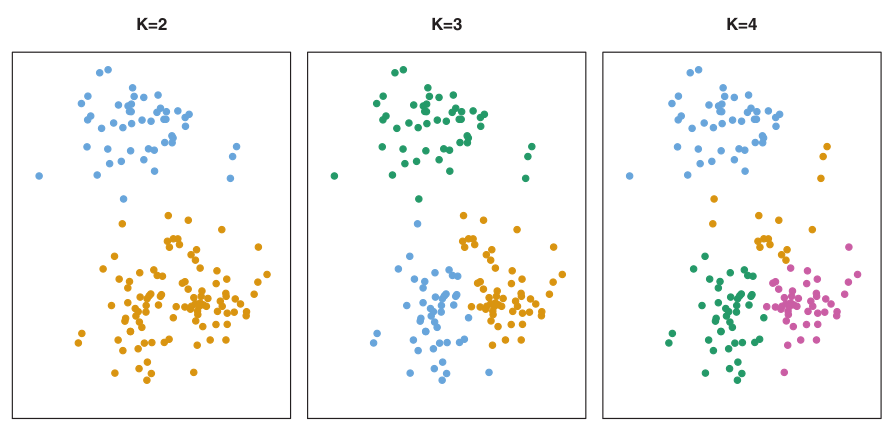
K-means clustering
- Simple approach to partitioning a data set into K distinct, non-overlapping clusters.
- K is specified beforehand.
James et al. 2013
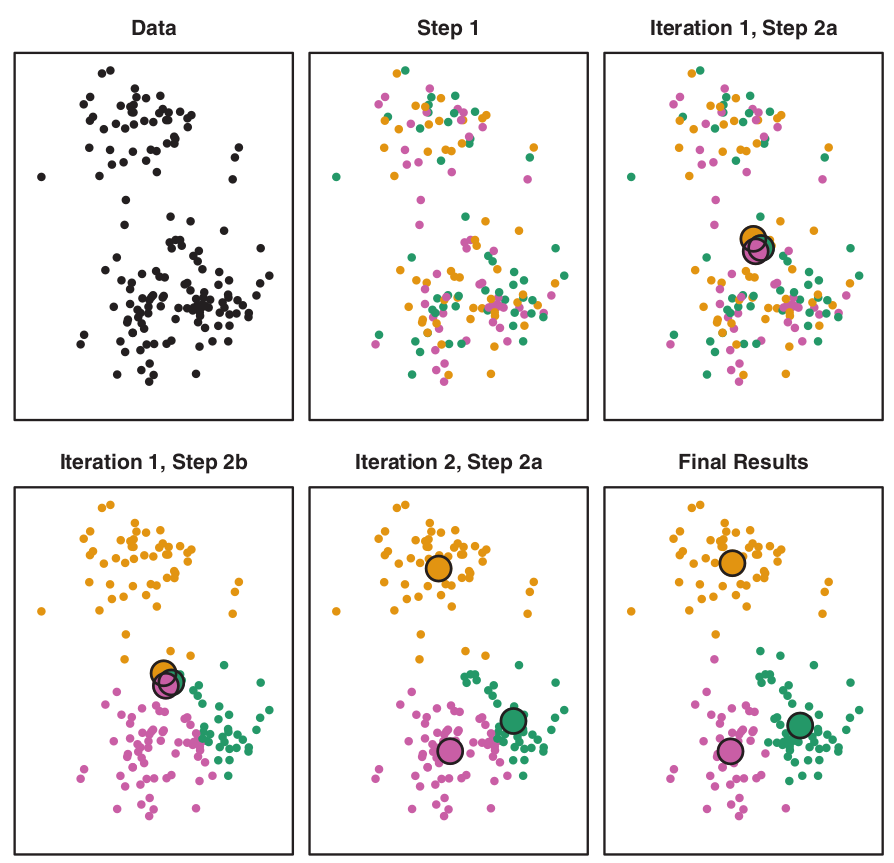
K-means algorithm
- \(K\) – number of clusters.
The process:
- Randomly assign each observation to groups 1 to K;
- For each of K clusters, derive its centroid (midpoint);
- Reassign each observation into a cluster whose centroid is the closest;
- Iterate until stability in cluster assignments is reached (local vs global optima).
Some properties of K-means partitioning
- Standard algorithm for k-means clustering is using Euclidean distance (distances are calculated using Pythagorean theorem).
- Different local optima (stability) can be reached.
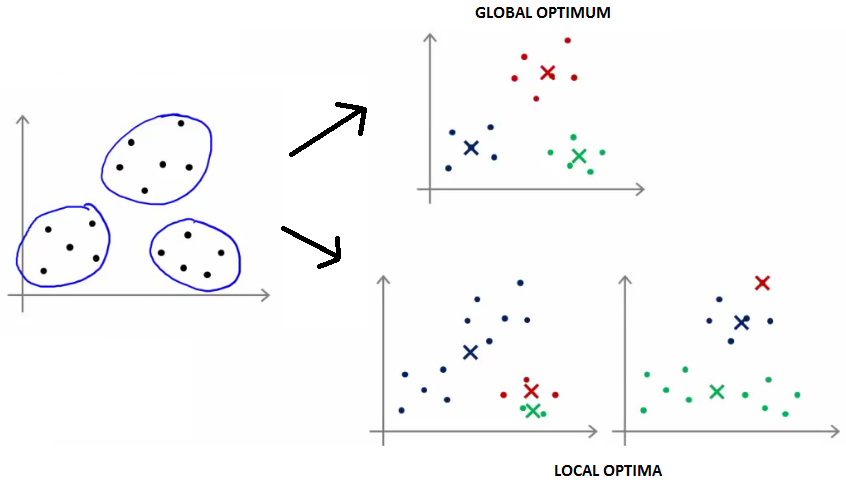
Number of clusters K must be specified in advance
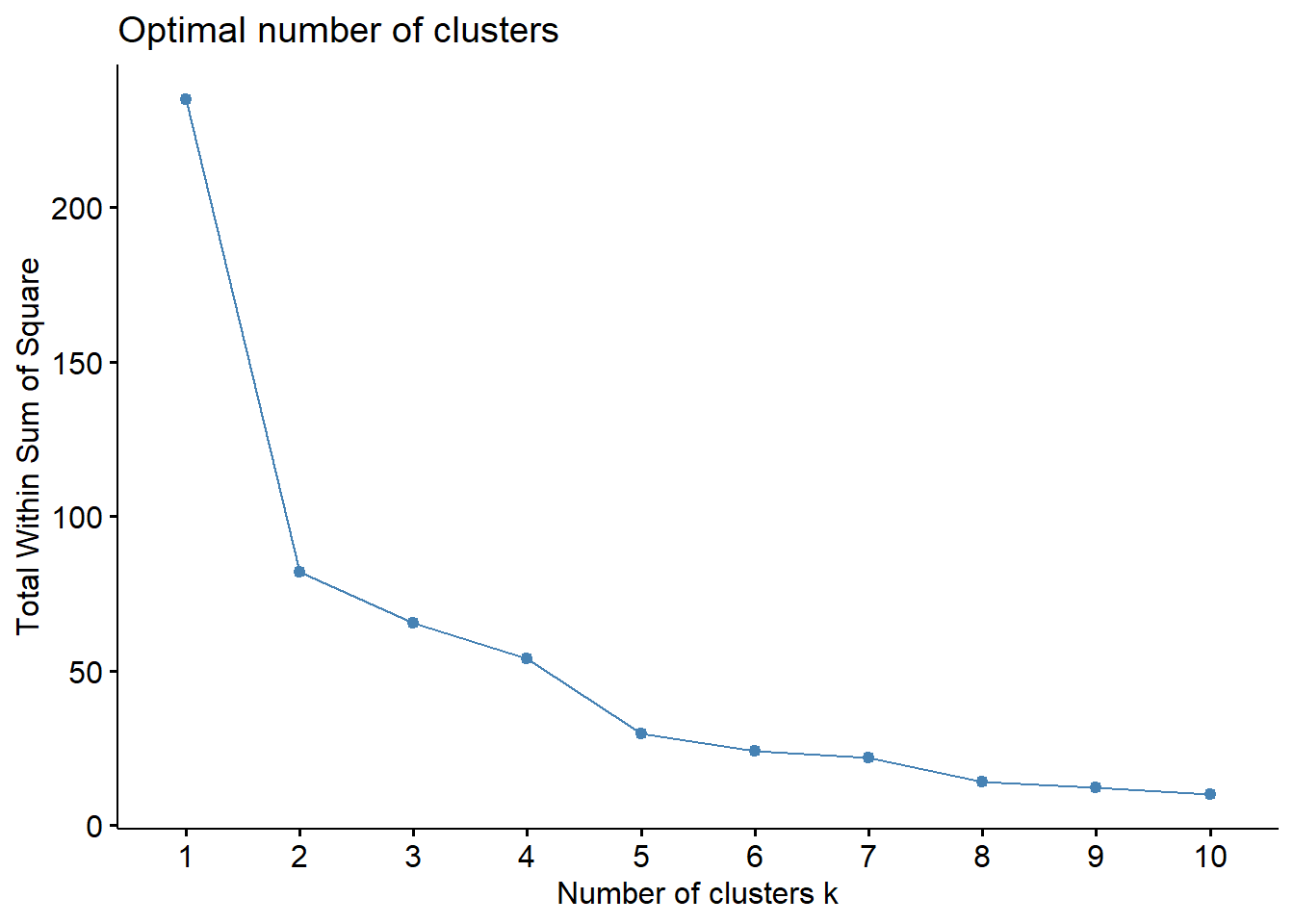
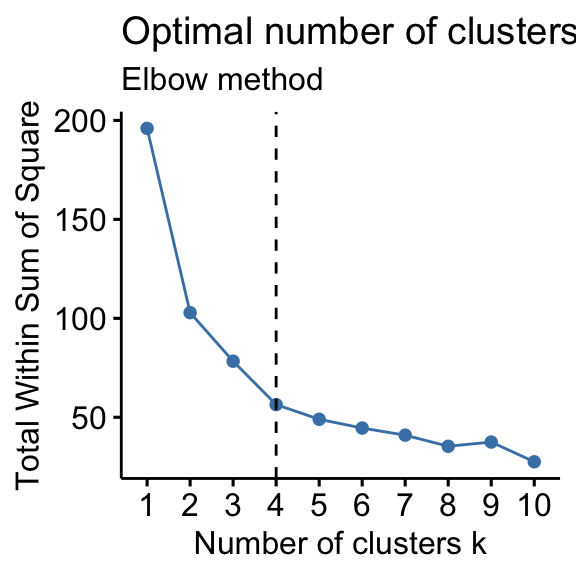
How to determine optimal number of clusters?
- Elbow method,
- Silhouette method
PCA
Name Length Width Thickness B.Width J.Width H.Length Weight
1 Darl 42.8 15.8 5.8 11.3 10.6 11.6 3.6
2 Darl 37.5 16.3 6.1 12.1 11.3 8.2 3.6
3 Darl 40.3 16.1 6.3 13.5 11.7 8.3 4.0
4 Darl 30.6 17.1 4.0 12.6 11.2 8.9 2.3 Name Length Width Thickness B.Width J.Width H.Length Weight
40 Pedernales 56.2 22.6 8.5 13.5 18.4 18.3 9.4
41 Pedernales 47.1 20.9 7.5 13.6 18.2 18.5 6.7
42 Pedernales 64.1 27.2 10.2 13.2 17.0 15.5 15.1
43 Pedernales 65.0 31.6 10.1 10.9 17.7 23.0 4.6Scatterplot
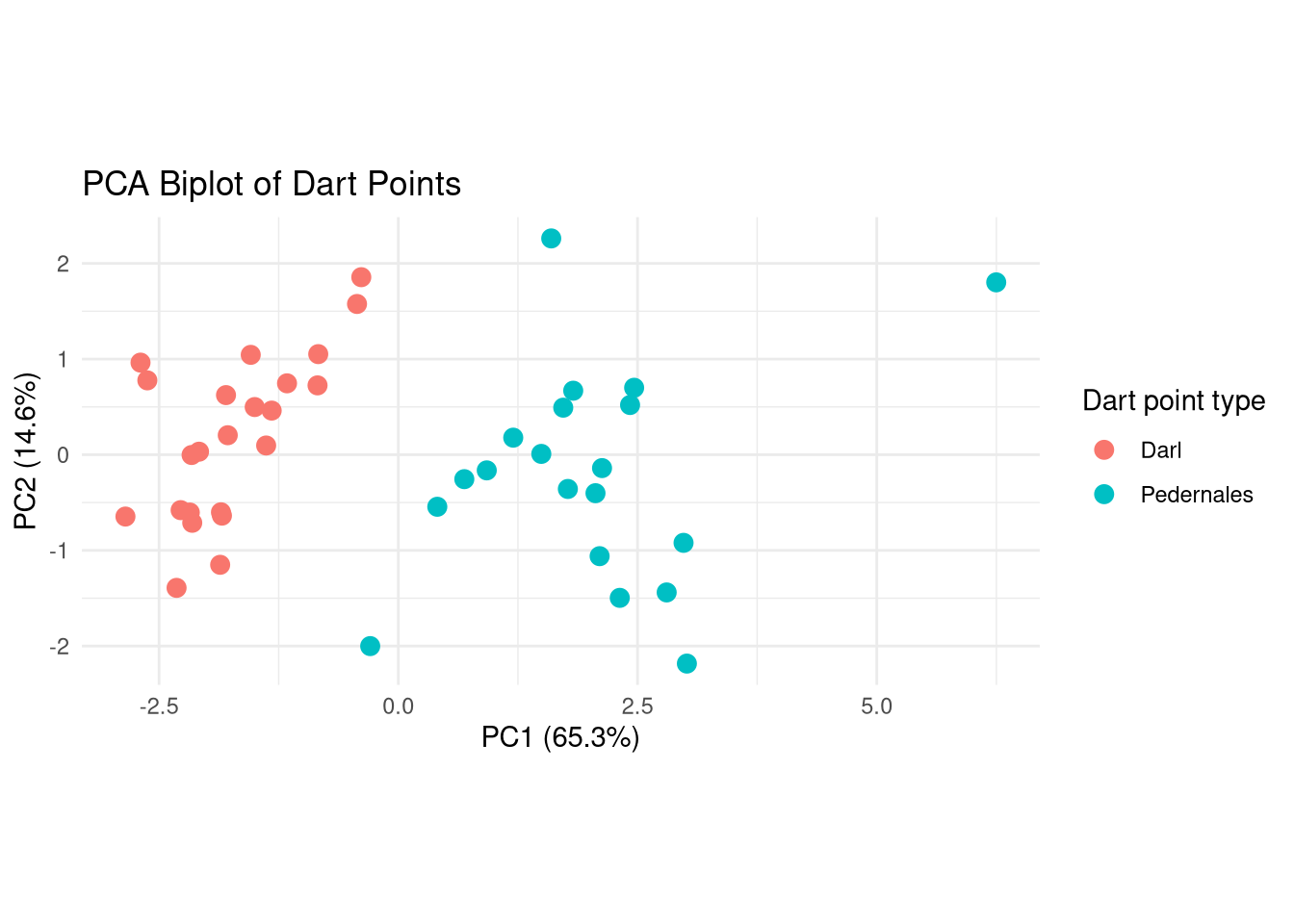
Reduce dimensions using PCA
Importance of components:
PC1 PC2 PC3 PC4 PC5 PC6 PC7
Standard deviation 2.1378 1.0104 0.80296 0.5898 0.46825 0.34354 0.28090
Proportion of Variance 0.6529 0.1459 0.09211 0.0497 0.03132 0.01686 0.01127
Cumulative Proportion 0.6529 0.7987 0.89085 0.9405 0.97187 0.98873 1.00000Biplot
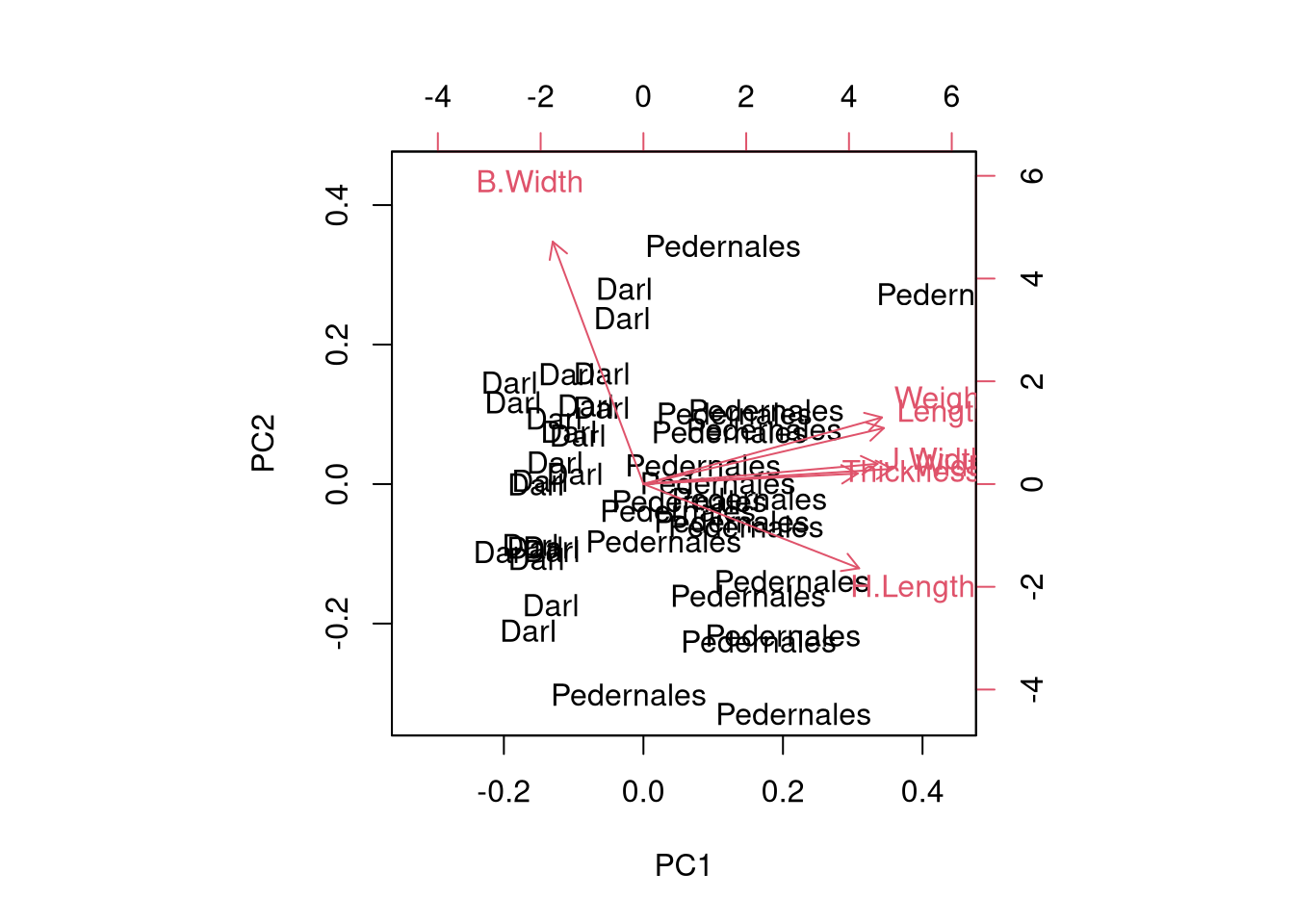
ggplot2 biplot
How many clusters?
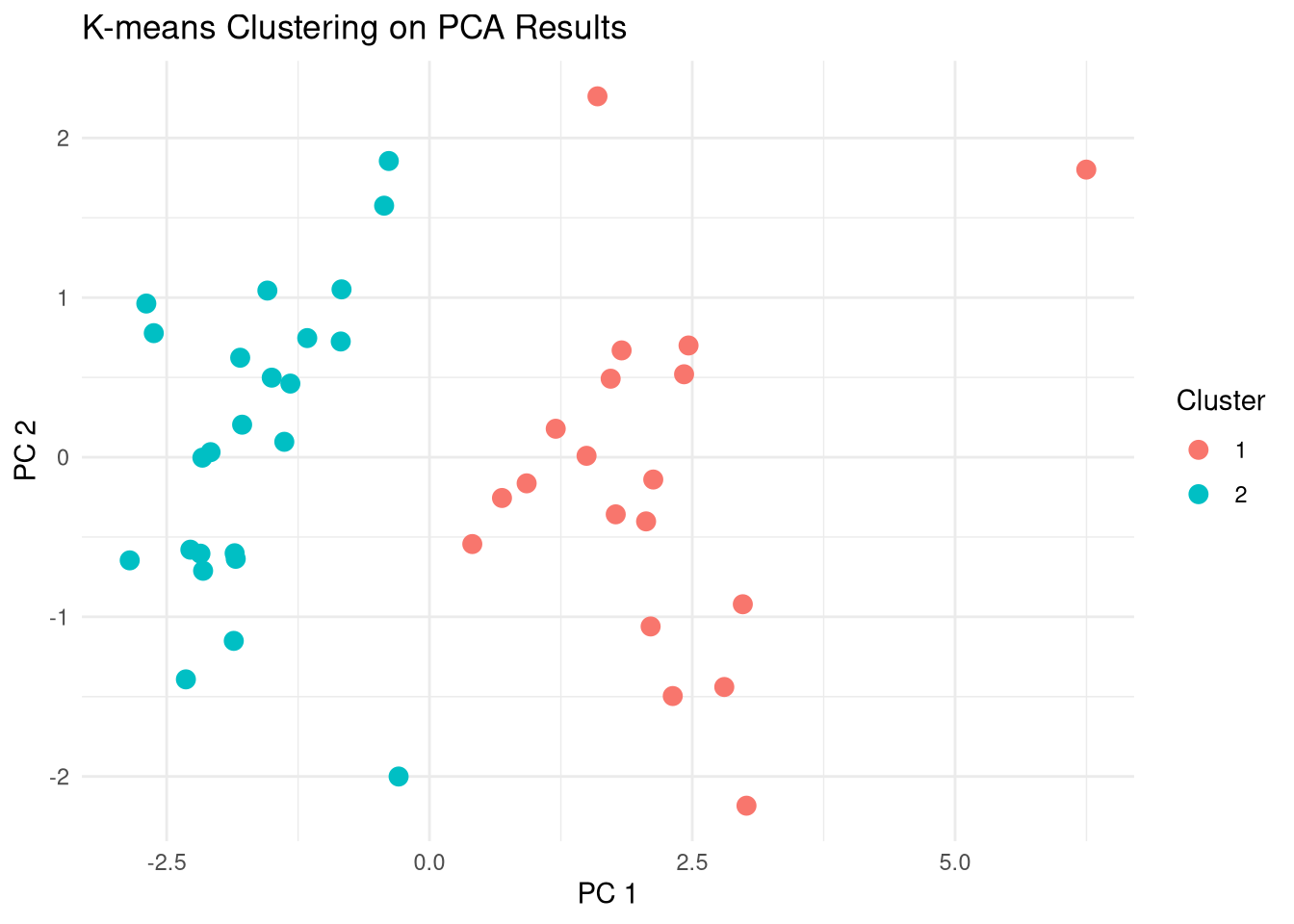
K-means
K-means clustering with 2 clusters of sizes 19, 24
Cluster means:
PC1 PC2 PC3 PC4 PC5 PC6
1 2.114733 -0.12266480 -0.1703675 0.02350401 -0.04361522 0.03322302
2 -1.674164 0.09710964 0.1348743 -0.01860734 0.03452871 -0.02630156
PC7
1 0.01907226
2 -0.01509887
Clustering vector:
Darl Darl.1 Darl.2 Darl.3 Darl.4
2 2 2 2 2
Darl.5 Darl.6 Darl.7 Darl.8 Darl.9
2 2 2 2 2
Darl.10 Darl.11 Darl.12 Darl.13 Darl.14
2 2 2 2 2
Darl.15 Darl.16 Darl.17 Darl.18 Darl.19
2 2 2 2 2
Darl.20 Darl.21 Darl.22 Pedernales Pedernales.1
2 2 2 1 1
Pedernales.2 Pedernales.3 Pedernales.4 Pedernales.5 Pedernales.6
1 1 1 1 2
Pedernales.7 Pedernales.8 Pedernales.9 Pedernales.10 Pedernales.11
1 1 1 1 1
Pedernales.12 Pedernales.13 Pedernales.14 Pedernales.15 Pedernales.16
1 1 1 1 1
Pedernales.17 Pedernales.18 Pedernales.19
1 1 1
Within cluster sum of squares by cluster:
[1] 88.53841 51.59016
(between_SS / total_SS = 52.3 %)
Available components:
[1] "cluster" "centers" "totss" "withinss" "tot.withinss"
[6] "betweenss" "size" "iter" "ifault" Plot
Compare clusters with Names
Darl Pedernales
1 0 19
2 23 1# Compare clusters to original names, i.e., dart point types
pca_scores |>
ggplot() +
aes(
x = PC1, y = PC2,
color = dp$Name,
shape = factor(kmeans_result$cluster)
) +
geom_point(size = 3) +
theme_minimal() +
labs(title = "K-means Clustering on PCA Results",
x = "PC 1",
y = "PC 2",
color = "Type of dartpoint",
shape = "Cluster")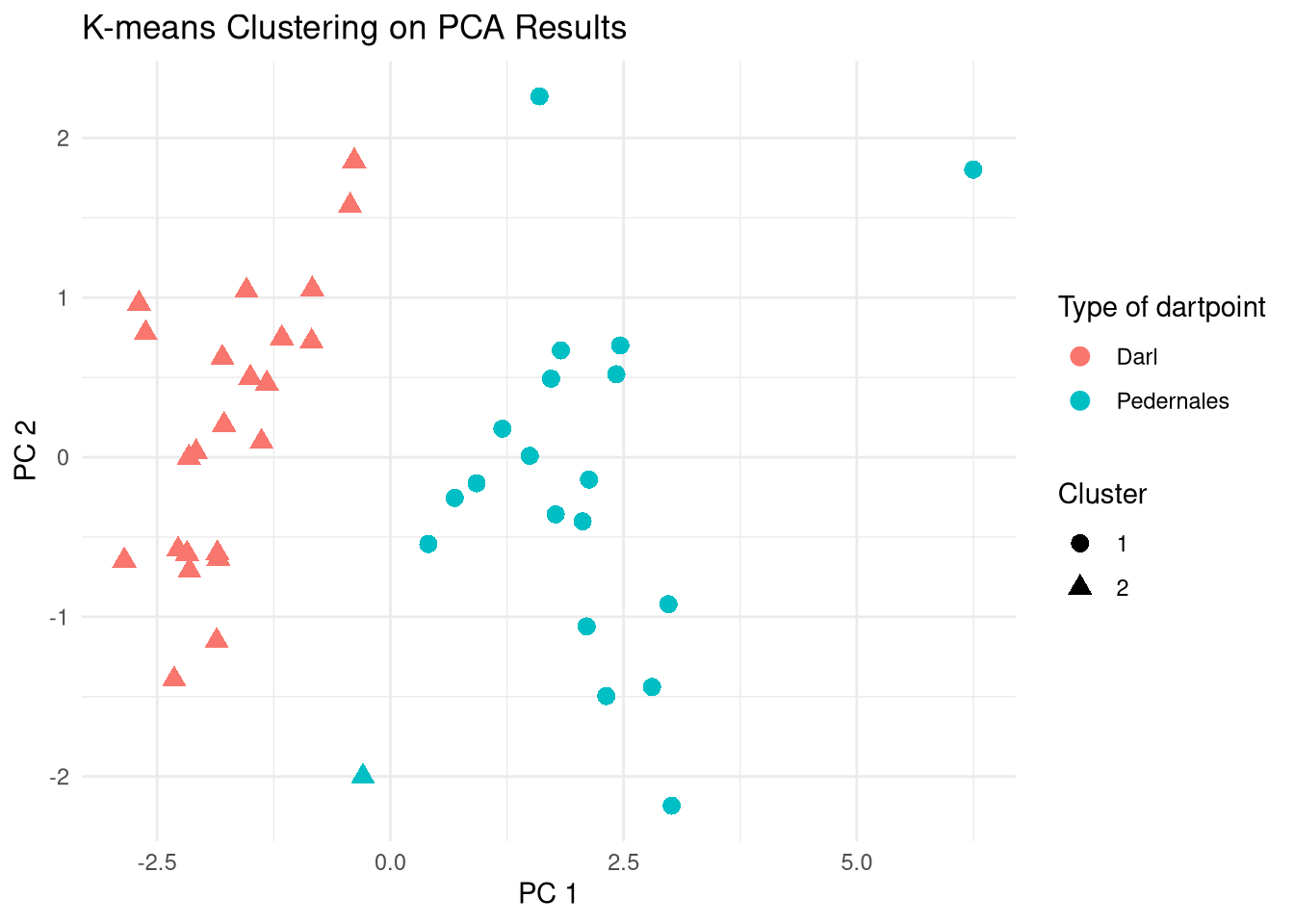
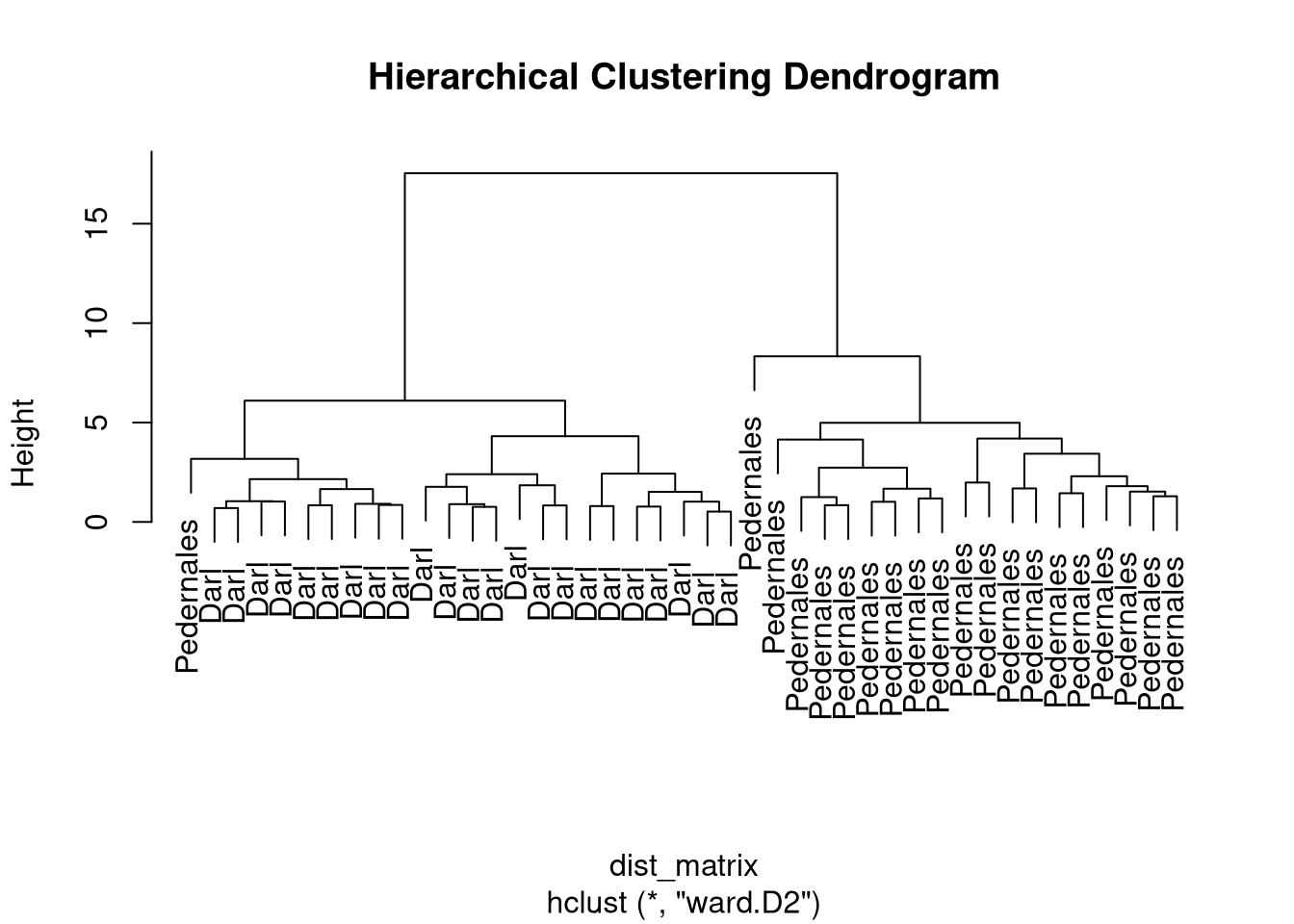
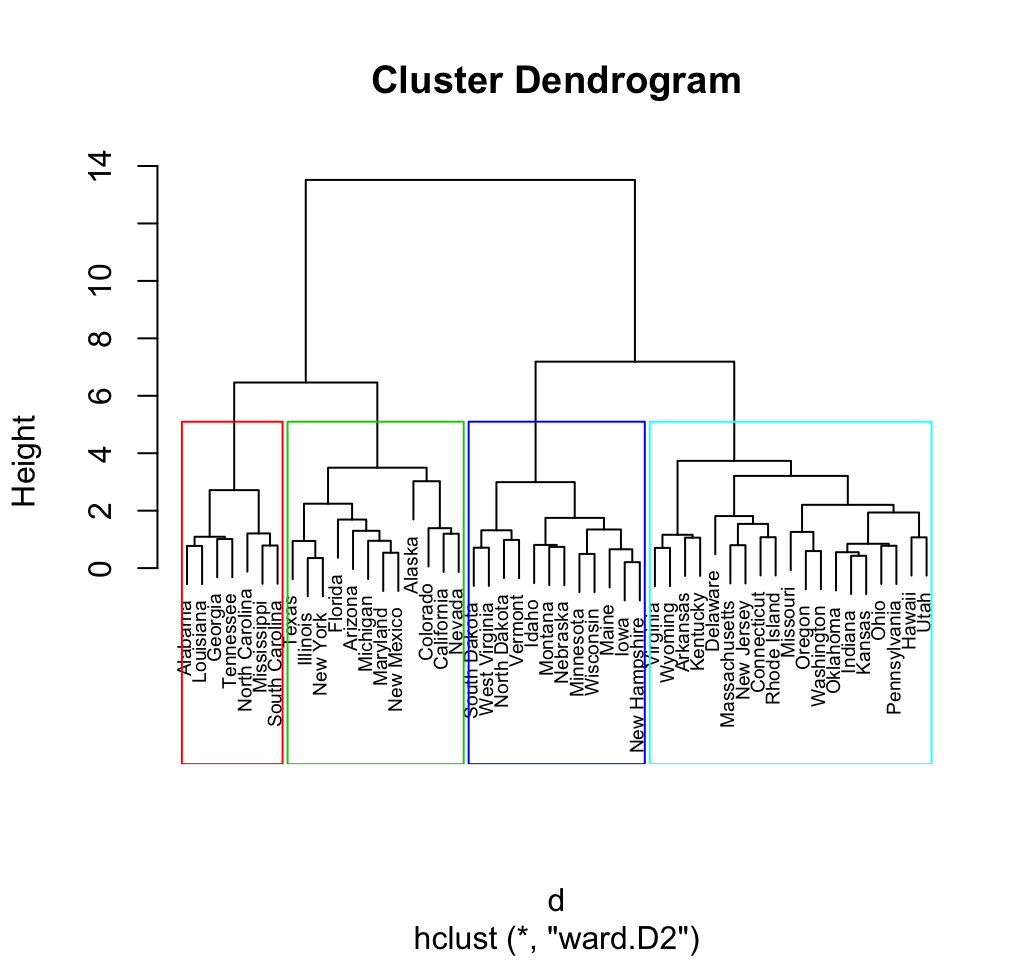
Hierarchical clustering
- The number of clusters is not specified beforehand.
- Output is a dendrogram – a hierarchical structure visualizing the cluster growth.
- Starts with a distance matrix.
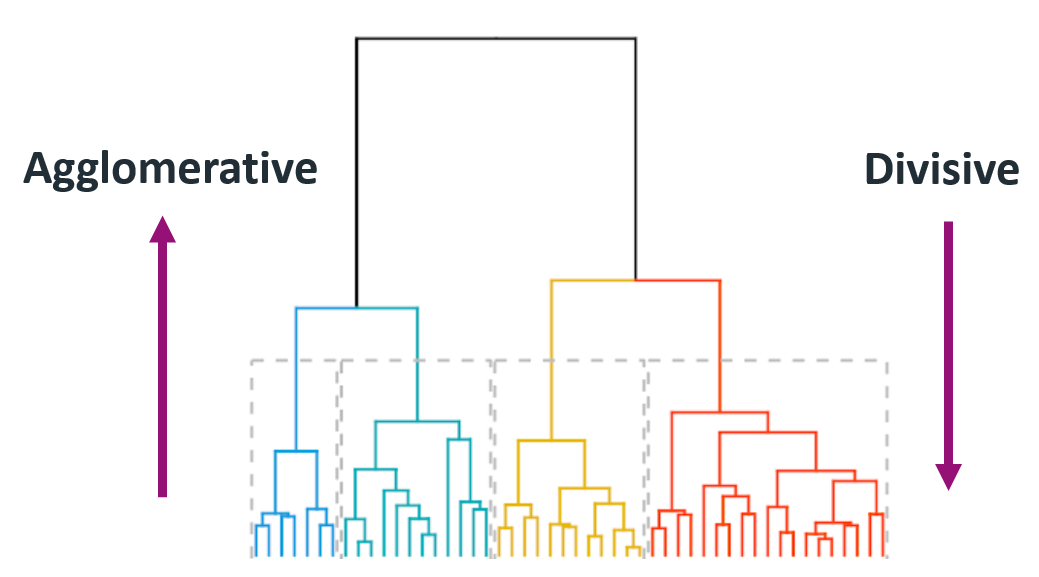
Agglomerative
- Builds the hierarchy of clusters from bottom-up until a single cluster is reached.
- Put each object in its own cluster;
- Join the clusters that are the closest;
- Iterate until a single cluster encompassing all objects is reached.
Divisive
- Divides a single large cluster into individual objects (top-down).
- Put all objects into a single cluster;
- Divide the cluster into subclusters at a similar distance;
- Iterate util all objects are in their own clusters.
Hierarchical clustering algorithms
Distance matrix
PC1 PC2 PC3 PC4 PC5
Darl -1.861665 -1.15057628 0.5968967 -0.3025608 0.56428007
Darl.1 -2.178930 -0.60404605 0.5255111 -0.7277931 -0.03876516
Darl.2 -2.082508 0.03183789 0.3072156 -0.6342330 0.07207282
Darl.3 -2.851549 -0.64616709 0.8645401 0.3607771 -0.24094772
Darl.4 -2.274738 -0.57920790 0.8686159 0.5521698 0.19617583 Darl Darl.1 Darl.2 Darl.3 Darl.4
Darl 0.000000 1.0151059 1.3907752 1.6191148 1.2428550
Darl.1 1.015106 0.0000000 0.6980987 1.3570655 1.4128283
Darl.2 1.390775 0.6980987 0.0000000 1.5855669 1.5079124
Darl.3 1.619115 1.3570655 1.5855669 0.0000000 0.8904547
Darl.4 1.242855 1.4128283 1.5079124 0.8904547 0.0000000Dendrogram
Clusters
- Different options where to cut the dendrogram…
- function
cutree(x, k, h) - specify
k– number of clusters or h– height where to cut the dendrogram
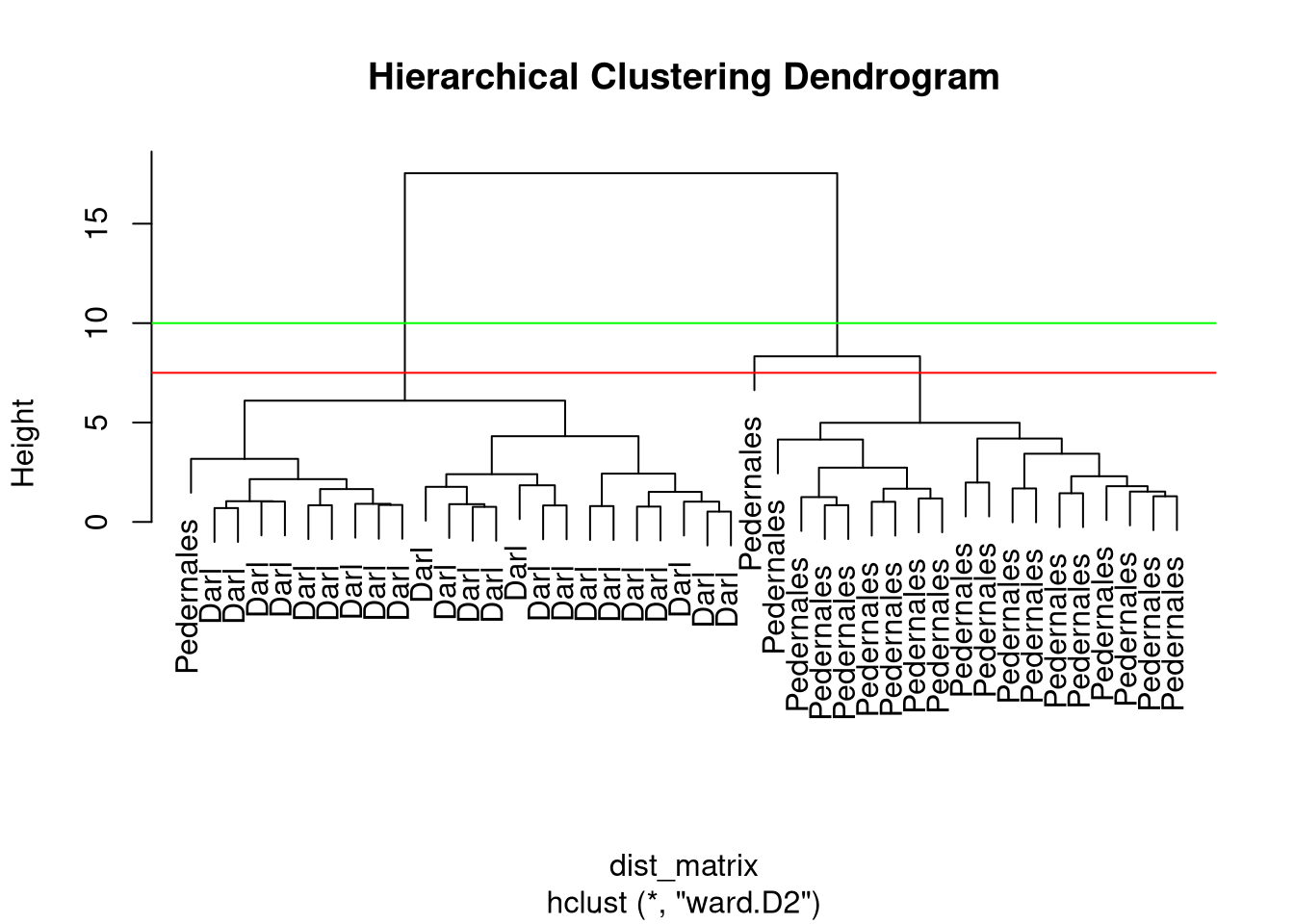
Cluster assignments
[1] 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 2 2 2 2 2 2 1 2 2 2 2 2 2 2 2
[39] 2 2 2 2 2Sidenote: Cluster linkage methods
Complete (maximum) linkage
- Largest distance between clusters.
method = "complete"
Single (minimum) linkage
- Smallest distance between clusters.
method = "single"
Mean (average) linkage
- Mean distance between clusters.
method = "average"
Wards method
- Minimizes total within cluster variance.
method = "ward.D2"
Clustering methods comparison
K-means partitioning
- Pre-specified number of clusters.
- Clusters may vary (different local optima).
- Best when groups in data are (hyper)spheres.

Hierarchical clustering
- Variable cluster numbers.
- Clusters are stable.
- Any shape of data distribution.

AES_707 Statistics seminar for archaeologists | Clustering